Researchers have developed a new technology that can accurately detect bacillary dysentery-causing bacteria and count how many of them there are, for the first time in the history of food and nutritional sciences.
The Rural Development Administration (RDA) announced recently it has developed a DNA probe that only joins with Shigella sonnei, a bacteria that causes bacillary dysentery, which is one of the main causes of food poisoning. Bacillary dysentery, or shigellosis, whose symptoms include fever, nausea, stomach cramps, vomiting and diarrhea, can be transmitted via contaminated water or food and only a medium or small number of bacteria- even 10 or 100 of them- can cause severe infection. It is very similar to pathogenic coliform bacillus and is thus difficult to detect.
In the past, the detection of bacteria that causes waterborne diseases used to be only possible through incubation, which can only be applied to food and patients. However, the new technology using a DNA probe can be done on various samples, such as water, soil and any surface area, and quickly detect pathogenic bacteria that might cause food poisoning.
The new detection method involves inserting and processing bacteria or DNA collected from water or food presumably contaminated with Shigella sonnei and inserting it into a quantitative polymerase chain reaction (qPCR) device, which is equipped with a DNA probe. The existence of a fluorescence reaction from the DNA probe can quickly determine whether or not an infection is present. By using this method, researchers can detect Shigella sonnei and find out the number of bacteria living in a certain area.
The new method can also drastically reduce the time needed to detect the bacteria, to only 30 minutes to one hour. The previous method required incubation of the bacteria samples for a day or more followed by the colony of bacteria needing to be separated for incubation for another one to three days to make it visible for accurate detection. The new method cuts the cost of detection by 30 percent, compared to the previous method.
The new method was published in the Journal of Microbiology and Biotechnology in August 2013.
“In December 2013, there were mass food poisoning incidents in Incheon and Gyeonggi-do (Gyeonggi Province) and there is a need for accurate and highly responsive detection technology,” said an RDA researcher. “With the development of this new method, it will now be possible to accurately detect bacteria from food and agricultural products. From now on, we will make more effort to develop DNA probes using genome analysis.”
By Limb Jae-un
Korea.net Staff Writer
jun2@korea.kr

The Rural Development Administration (RDA) announced recently it has developed a DNA probe that only joins with Shigella sonnei, a bacteria that causes bacillary dysentery, which is one of the main causes of food poisoning. Bacillary dysentery, or shigellosis, whose symptoms include fever, nausea, stomach cramps, vomiting and diarrhea, can be transmitted via contaminated water or food and only a medium or small number of bacteria- even 10 or 100 of them- can cause severe infection. It is very similar to pathogenic coliform bacillus and is thus difficult to detect.

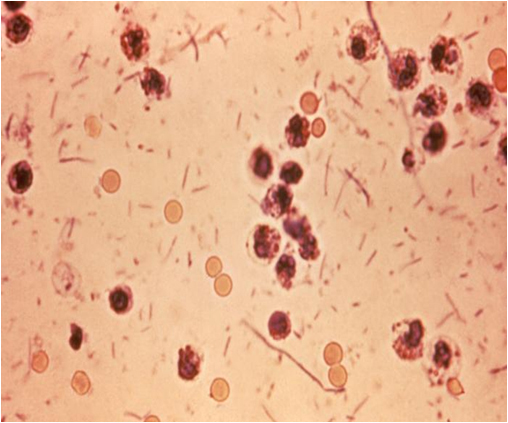
A magnified view of Shigella sonnei as seen through a microscope (Photo courtesy of the RDA)

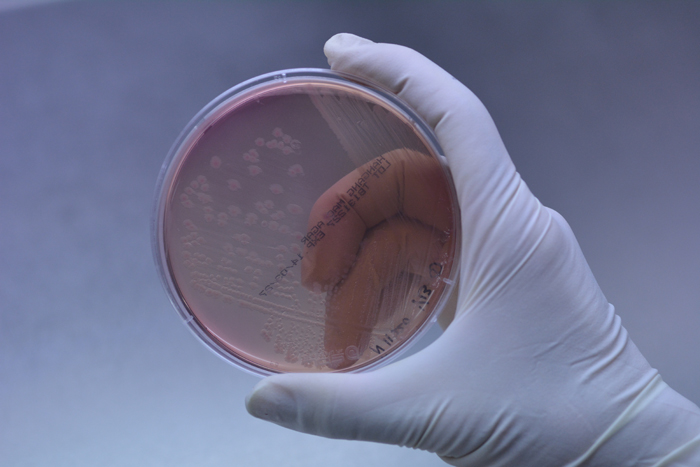
Shigella sonnei bacteria cultivated in a Petri dish (Photo courtesy of the RDA)
In the past, the detection of bacteria that causes waterborne diseases used to be only possible through incubation, which can only be applied to food and patients. However, the new technology using a DNA probe can be done on various samples, such as water, soil and any surface area, and quickly detect pathogenic bacteria that might cause food poisoning.
The new detection method involves inserting and processing bacteria or DNA collected from water or food presumably contaminated with Shigella sonnei and inserting it into a quantitative polymerase chain reaction (qPCR) device, which is equipped with a DNA probe. The existence of a fluorescence reaction from the DNA probe can quickly determine whether or not an infection is present. By using this method, researchers can detect Shigella sonnei and find out the number of bacteria living in a certain area.
The new method can also drastically reduce the time needed to detect the bacteria, to only 30 minutes to one hour. The previous method required incubation of the bacteria samples for a day or more followed by the colony of bacteria needing to be separated for incubation for another one to three days to make it visible for accurate detection. The new method cuts the cost of detection by 30 percent, compared to the previous method.

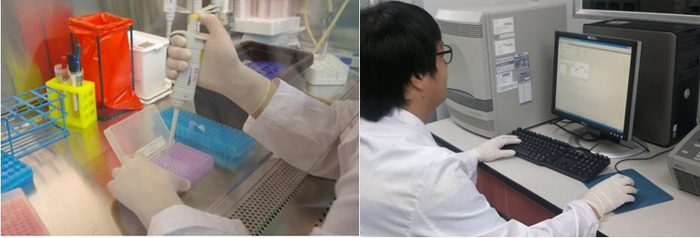
DNA analysis is being carried out to detect bacillary dysentery-causing bacteria. (Photo courtesy of the RDA)
The new method was published in the Journal of Microbiology and Biotechnology in August 2013.
“In December 2013, there were mass food poisoning incidents in Incheon and Gyeonggi-do (Gyeonggi Province) and there is a need for accurate and highly responsive detection technology,” said an RDA researcher. “With the development of this new method, it will now be possible to accurately detect bacteria from food and agricultural products. From now on, we will make more effort to develop DNA probes using genome analysis.”
By Limb Jae-un
Korea.net Staff Writer
jun2@korea.kr
